
SciMethods: Paired Blind Polytrode and Juxtacellular Recordings In Vivo
Joana P. Neto1, Adam R. Kampffsup1 & Christian Wilms2
1 Champalimaud Centre for the Unknown, Lisbon, Portugal
2 Scientifica Ltd. Uckfield, UK
Introduction
Simultaneous recordings from multiple separate electrodes in close proximity (<100 μm) are an extremely versatile experimental configuration. In the past this approach has been used to record from multiple sites of a single neuron1, multiple neurons in close proximity2 and to correlate intracellular activity with extracellular polytrode recordings3, amongst others. By far the majority of studies using this configuration have been performed in slice preparations. In contrast, in vivo use of simultaneous recordings from multiple closely spaced sites has remained elusive with only a handful of research groups performing such experiments4,5. The reason for this is the disproportionate increase in difficulty of bringing the undamaged electrodes into close proximity to each other when switching from in vitro to in vivo settings. While it is easily possible to visualise pipette positions in slices using brightfield microscopy, this is not an option in the intact brain. There are two approaches to circumventing this problem: The first is to use a two-photon imaging system to visualise the electrodes in deeper tissue. This method has been used to perform whole-cell recordings from up to four cells simultaneously4, but is expensive, limited to depths of 1 mm at best and requires that the electrodes fluoresce. The second approach is to combine computer controllable motorised micromanipulators with trigonometric calculations of the electrode positions in the tissue. This method is cost efficient and not limited to the most superficial brain layers, but in practice, it has rarely been used, as it has been difficult to implement reliably. In this technical note, we present a recent advance in the latter approach6. We present this straightforward algorithm to achieve blind ‘paired recordings’ in a reliable manner using Scientifica micromanipulators.
Implementation
The required components are listed below. The specific models used for this application note are listed in italics.
Hardware
- Two motorised manipulators (Scientifica IVM and PatchStar)
- Upright microscope with a large working distance and oblique illumination for visualising the electrode positions above the brain surface (custom built)
Software
- Software for calibration and control of manipulators, video monitoring and voltage recording (custom software implemented using Bonsai).
Description
The manipulators were positioned on each side of the stereotactic frame so that they were exactly opposite each other (see Figure 1A). Next, the two electrode axes were set to the respective desired angles: The juxtacellular electrode (mounted on the PatchStar) was set to 61º from horizontal, while the polytrode (on the IVM) was set to -48.2º from horizontal. The PatchStar allows the ‘approach’ angle of the electrode to be freely adjusted, combining the movement of the X- and Z-axes to move in a straight diagonal line along the set angle of the electrode holder. The IVM does not and thus needs to be tilted around the Y-axis as a whole.
To allow software calibration and precise electrode positioning the manipulators have to be perfectly parallel (Figure 1B). To achieve this, the manipulators are mechanically aligned using a machinist’s dial. In the first step, the PatchStar was squared to the stereotactic frame. Once achieved, the IVM was aligned to ensure it was square to the frame and thus also parallel to the PatchStar.
Next, the two manipulators need to be brought into a common coordinate system in order for both electrodes to be moved in unambiguous reference to each other. As both are set up for different approach angles, these angles need to be taken into consideration. To achieve this, a coordinate transformation must be applied to one of them (in our case the IVM). Instead of calculating this transform using basic trigonometry, a transformation matrix (or ‘look up table’) is determined by direct measurement. A long working distance microscope was set up to image electrode positions in the XZ and/or XY planes (Figure 1B, C) and the coordinates were set to zero (X=0, Y=0, Z=0). Next, the juxtacellular electrode (PatchStar) was moved to a point in the space, and the microscope was moved and refocused to recentre the tip of the pipette in the crosshair. Then the extracellular electrode (IVM) was moved to the same point. This process was repeated for several different locations (n = 15) that spanned a large volume (5000 x 5000 x 5000 μm, in 1000 μm steps) and the coordinates for both electrodes were recorded. The resulting pairing at each point was then used to generate the transformation matrix. This calibration procedure, as well as the coordinate transformation, was implemented in Bonsai, a free open-source visual programming environment (https://bitbucket.org/horizongir/bonsai; Reference 7).
Following the mechanical alignment and software calibration, the error in the distance estimation can be reduced to the level of 10.5 ± 5.2 μm, roughly the size of a neuronal cell body.
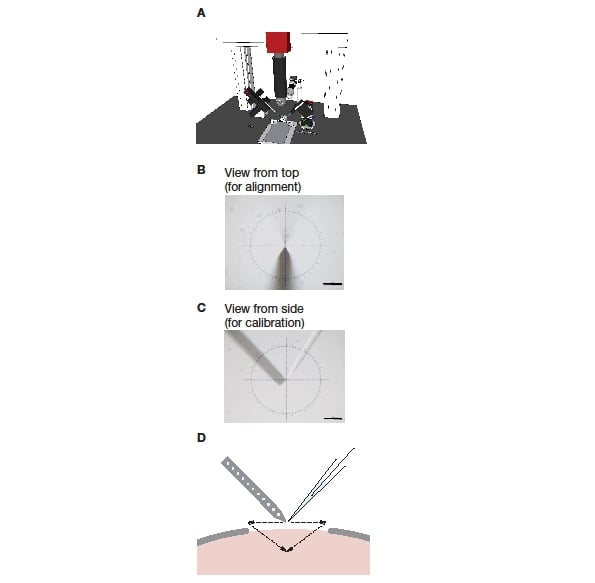
Figure 1: Setup of a blind dual recording system. A: The system consisting of a stereotactic frame, two micromanipulators, and a long working distance microscope mounted on a frame. B: View onto the two electrodes from the top, as used during mechanical alignment. C: View onto the two electrodes from the side, as used during software calibration. D: Schematic of pipette positioning and insertion during an experiment.
Running an experiment
At the start of a recording, the two electrodes are sequentially positioned at the centre of the microscope image and this position is set to ‘zero’ for each electrode. This registration should be performed directly above the intended recording location and as close as possible to the dura. While a microelectrode has a clear origin point (the tip), on a polytrode, any point can be selected as origin, simply by positioning that point in the centre of the image for zeroing. After setting the origins of both electrodes, the microscope is no longer required for positioning.
From this point on, the electrodes can be positioned relative to each other, within the precision achieved in the initial alignment and calibration steps. If visually guided insertion of the electrodes is desired, the microscope can now be replaced by a macrozoom lens.
After ‘zeroing’, each electrode is retracted and positioned so that it can be advanced into the final position using mainly movement along the electrode axis (Figure 1D). This reduces the amount of damage to the tissue.
Ideally, the larger of the two electrodes (in our experiments the polytrode) is inserted into the brain and brought to the final recording position first. This helps avoid movement of the tissue after positioning the first electrode.
Conclusion
In this technical note, we have briefly presented a straightforward approach to achieving blind paired recordings in vivo. A detailed discussion along with videos of the process can be found in Reference 6.
PatchStar Micromanipulator
The most versatile motorised manipulator for electrophysiological studies. Designed with leading physiologists, the PatchStar is also perfect for microinjection and other demanding positioning requirements.
References
1. Stuart, G. & Hausser, M. Initiation and spread of sodium action potentials in cerebellar Purkinje cells. Neuron 13, 703–712 (1994).
2. Ko, H. et al. Functional specificity of local synaptic connections in neocortical networks. Nature 473, 87–91 (2011).
3. Anastassiou, C. A., Perin, R., Buzsáki, G., Markram, H. & Koch, C. Cell type- and activity-dependent extracellular correlates of intracellular spiking. J. Neurophysiol. 114, 608–623 (2015).
4. Jouhanneau, J.-S., Kremkow, J., Dorrn, A. L. & Poulet, J. F. A. In Vivo Monosynaptic Excitatory Transmission between Layer 2 Cortical Pyramidal Neurons. Cell Rep. 13, 2098–2106 (2015).
5. London, M., Roth, A., Beeren, L., Häusser, M. & Latham, P. E. Sensitivity to perturbations in vivo implies high noise and suggests rate coding in cortex. Nature 466, 123–127 (2010)
6. Neto, J. P. et al. Validating silicon polytrodes with paired juxtacellular recordings: method and dataset. J. Neurophysiol. 116, (2016).
7. Lopes, G. et al. Bonsai: an event-based framework for processing and controlling data streams. Front. Neuroinform. 9, 7 (2015).
Other SciMethods you may be interested in...
SciMethods: Simultaneously recording from four neurons using PatchStar and MicroStar manipulators
SciMethods: Visual Stimulation of Retinal Explants on a Standard Multiphoton Microscope
SciMethods: Using a Scientifica Multiphoton SliceScope for Fluorescence Lifetime Imaging
SciMethods: Controlling and Coordinating Experiments in Neurophysiology


)