#LabHacks: Choosing the best opsin for your optogenetics experiments
According to Tye and Deisseroth 20121, designing an optogenetics experiment can be broken down into five main parts, the first of which is finding the most suitable opsin for your particular investigation.
Since the establishment of optogenetics as a powerful technique to study neural circuit function, several research groups have worked on expanding the number of available opsins. As such the “optogenetics toolbox” has grown significantly, magnifying the number of processes that can be interrogated, but complicating the selection process.
Deciding which opsin to use requires the consideration of several trade-offs, while always keeping the experimental goals in mind. These include – but are not limited to – ion conductance, peak activation wavelength, light sensitivity and kinetics.
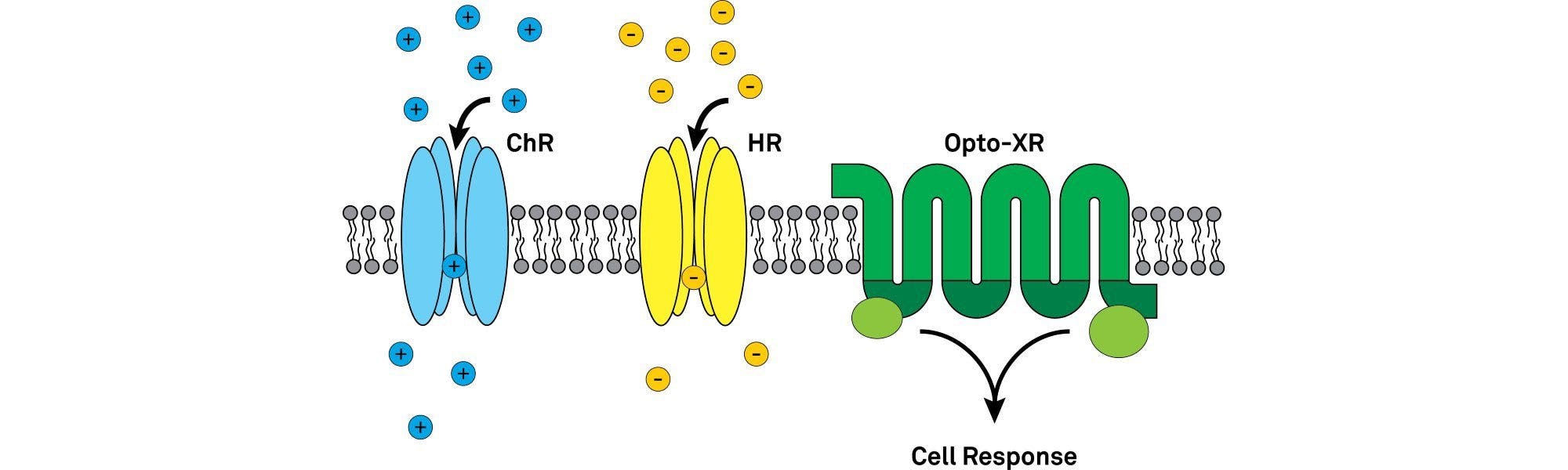
The opsins used in optogenetics can be widely divided into type I (microbial opsins) and II (animal/vertebrate opsins)2. The microbial opsins first used in optogenetic photomanipulation of neural tissues and include the widely-known channelrhodopsin, halorhodopsin and bacteriorhodopsin families. They are single component light sensing systems (i.e. the same protein confers light sensitivity and ion conductance). Type I opsins work by directly altering the membrane potential of the neurons.
Type II opsins are mainly comprised of vertebrate rhodopsins, like the light-sensing proteins that enable you to see. They are light-sensitive G-protein coupled receptors (GPCRs). When activated by light a conformational change in the protein’s structure starts an intracellular signalling cascade which can ultimately alter the inhibitory or excitatory state within the cell.
The microbial opsins
Microbial opsins offer fast and precise manipulation of neuronal excitability by directly altering the membrane potential of the neuron to bring it closer to, above, or further away from the threshold for generating action potentials.
Fast excitation
Originating from microalgae of the genus Chlamydomonas, the channelrhodopsins are light-gated ion channels3. Boyden et al. 20054 used Channelrhodopsin-2 (ChR2) to show optical control of neural activity in their famous Nature Neuroscience paper.
ChR2 is a cation channel that leads to rapid depolarization of a neuron by enabling cations (e.g. Na+, K+, Ca2+, H+) to move into the cell (in the direction of the electrochemical gradient), raising the membrane potential and evoking precisely timed action potentials5. ChR2 has a peak activation wavelength of 470 nm (blue light) and decay kinetics of ~10 ms at room temperature2.
A variety of other rapidly depolarising optogenetic opsins have been developed from ChR2 mutations, channelrhodopsins from other species and the creation of chimaeras from combinations of channelrhodopsins. This has led to tools with slower kinetics, shifted wavelengths, higher sensitivity, etc. For a list of the properties on several channelrhodopsins read Yizhar et al. 20112.
One example is humanised ChR2 (hChR2), a widely-used opsin whose genetic code replaced algal codons with mammalian codons leading to better expression levels in mammalian animal models.
Fast inhibition
Bacteriorhodopsins (BR), proteorhodopsins (PR) and archeorhosopsins (AR) are all inhibitory proton pumps that hyperpolarize neurons by pumping H+ out of the cell. This leads to a more negative membrane potential making excitatory inputs into the cell less likely to reach the action potential threshold. Several naturally occurring light-sensitive proton pumps have been identified or engineered for slightly differing properties like enhanced expression on the cellular membrane, altered kinetics and different peak activation wavelengths.
Halorhodopsin (NpHR), so called after the archaeon where it naturally occurs (Natronomonas pharaonic), is another inhibitory opsin that pumps chloride ions into the cell to hyperpolarize the membrane6. The mammalian codon optimised form of this opsin did not traffic to the outer cell membrane and accumulated in the endoplasmic reticulum. Further engineering led to eNpHR3.0 which induces a large photocurrent and localises to the outer membrane. This is now among the most commonly used inhibitory opsins and is often used in combination with an excitatory opsin to test individual circuit components7. Its peak activation wavelength of 590 nm (yellow light) is far enough from the commonly used ChR2 wavelengths (470 nm, blue light) to avoid involuntary cross-activation.
The animal opsins
Control of intracellular signalling
Engineered versions of vertebrate rhodopsins have been developed to take advantage of their light-activated nature and initiation of intracellular signalling pathways. The most common group of these, known as optoXRs, are opsin-receptor chimaeras of a rhodopsin and conventional ligand-gate GPCR8. In naming each member of the family, the X is replaced by the GPCR being used (e.g. Optoα1AR for a rhodopsin and α1 adrenergic receptor chimaera)9. They allow optical modulation of a variety of intracellular signalling cascades with temporally precise resolution suitable for studying behaviour in freely moving mice.
Bistable Opsins
Step function opsins (SFOs) are a family of ChR2 mutants that once activated by light, show prolonged activity after termination of the light stimulus10. The first member of the group was ChR2(C128S), so called because it’s 128th residue was altered from a cysteine to a serine, remains activated for approximately 1.7 minutes when activated by 470 nm light. They can also be inactivated by light of a different wavelength, 560 nm peak inactivation wavelength in the case of ChR2(C128S). The ability of the opsins to remain activated for extended periods even after cessation of a light pulse, and to be turned off make them useful for experimental paradigms precise modification of the firing rate of neuronal populations.
Combining opsins
More complex experimental protocols can be developed with the introduction of different opsins into different cell types. In these cases, you will need to contemplate targeted expression specificity, the overlap of activation wavelengths, and whether light-activation of each opsin will occur in the same or different regions. Each of these factors may influence which opsin you choose.
In vitro, in vivo, light sensitivity and phototoxicity
Further considerations when choosing your opsins depend on what type of experiment you will be conducting. For example, different characteristics will be essential for in vitro network dynamics or in vivo behavioural experiments.
For in vitro experiments the peak activation wavelength of the opsin might not be the most important concern. You won’t necessarily be trying to penetrate deeply into thick, highly scattering tissue, so whether your opsin is best suited to a blue, yellow or red-shifted light source won’t affect performance too much. However, fast kinetics might be crucial for your experimental goals.
For behavioural experiments, the kinetics may be less important whereas light sensitivity may be crucial due to the power you need to achieve in the desired region and potential off-target effects caused by using too much light.
References
- Tye, K. M. & Deisseroth, K. Optogenetic investigation of neural circuits underlying brain disease in animal models. Nat Rev Neurosci 13, 251–266 (2012).
- Yizhar, O., Fenno, L. E., Davidson, T. J., Mogri, M. & Deisseroth, K. Optogenetics in Neural Systems. Neuron 71, 9–34 (2011).
- Channelrhodopsins - OpenOptogenetics.org. Available at: http://www.openoptogenetics.org/index.php?title=Channelrhodopsins. (Accessed: 8th March 2017)
- Boyden, E. S., Zhang, F., Bamberg, E., Nagel, G. & Deisseroth, K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci 8, 1263–1268 (2005).
- Mattis, J. et al. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nat Meth 9, 159–172 (2012).
- Guru, A., Post, R. J., Ho, Y.-Y. & Warden, M. R. Making Sense of Optogenetics. Int J Neuropsychopharmacol 18, (2015).
- Han, X. & Boyden, E. S. Multiple-Color Optical Activation, Silencing, and Desynchronization of Neural Activity, with Single-Spike Temporal Resolution. PLoS ONE 2, (2007).
- Airan, R. D., Thompson, K. R., Fenno, L. E., Bernstein, H. & Deisseroth, K. Temporally precise in vivo control of intracellular signalling. Nature 458, 1025–1029 (2009).
- Opto-XRs - OpenOptogenetics.org. Available at: http://www.openoptogenetics.org/index.php?title=Opto-XRs. (Accessed: 8th March 2017)
- Fenno, L., Yizhar, O. & Deisseroth, K. The development and application of optogenetics. Annu. Rev. Neurosci. 34, 389–412 (2011).
Banner Image Credit: Wikimedia Commons
Take a look at other #LabHacks articles
- #LabHacks: Tips for improving your electrophysiology experiments
- #LabHacks: How to align your laser for two-photon imaging
- #LabHacks: Tips for cleaning the optics of your microscope
- #LabHacks: To compensate or not to compensate, that is the question
- #LabHacks: How to reduce the noise around your electrophysiology rig

)