
New to two-photon imaging combined with optogenetics? This is what I learnt from my “failed” PhD project.

by Filipe Machado Ferreira, Research Associate, Khan Lab, KCL
Introduction
The intricate workings of the human brain bear striking resemblance to the harmonious coordination of an orchestra in full performance. Just as a conductor directs a symphony by stressing the tempo and the dynamics of soundwaves, the brain orchestrates spatiotemporal precise patterns of neural activity that shape what we see, feel, and identify with. This precise rhythmic composition is shaped by the interplay of different group of neurons with distinct morphological and physiological characteristics across multiple brain regions, just like different instruments within the strings and woodwind families play together to produce a beautiful well-balanced melody. Understanding how different neurons play together is the fundamental aspect of neuroscience research. In the last twenty years in vivo calcium imaging and optogenetic have revolutionised the way one studies neural circuits. While the first allows an indirect measurement of neural activity at different spatial scales, ranging from single spines to large areas of the cortex in rodents, the latter provides means to precisely stimulate or inhibit activity of single or large ensembles of neurons. This harmonious combination of targeted perturbations and dynamic readout of brain activity gives one the power to conduct precise changes in the melodies being played.
This romanticised view of all-optical approaches draws people’s attention and may even be considered “sexy” within the scientific community, and rightly so I would argue, as these tools are a product of great ingenuity and provide extremely valuable cutting-edge technology for unveiling the mystery behind how the brain works. However, several limiting aspects of all-optical approaches are not talked about enough and should be central in the experimental design of a research project. Here, I highlight some of the well-known technical limitations associated with the use of all-optical approaches, putting emphasis on issues I came across while navigating through failed experiments using optogenetics during my PhD. I also comment on how such limitations should be considered when interpreting the data collected and suggest (try to suggest) ways to tackle them.
Figure 1: Artificial intelligence generated image based on the first few lines of this article. Source: DeepAI. Input: ‘Just as a conductor directs a symphony by stressing the tempo and the dynamics of sound waves, the brain orchestrates spatiotemporal precise patterns of neural activity that shape what we see, feel, and identify with. This precise rhythmic composition is shaped by the interplay of different group of neurons with distinct morphological and physiological characteristics across multiple brain regions, just like different instruments within the strings and woodwind families play together to produce a beautiful well-balanced melody.’
Two-photon calcium imaging’s poor temporal resolution
In vivo calcium imaging is known to have poor temporal resolution when compared with electrophysiology recordings, as an all-optical experimental approach always faces a trade-off between temporal and spatial resolution, for hardware reasons. Moreover, although faster and brighter new calcium indicators are engineered to better accommodate one’s experimental needs, currently these still fall short on providing high-temporal resolution information from a large population of neurons. This impacts the ability to capture fast events, and thus is known to miss several single action potentials across the population. Therefore, deciding whether this loss of information is relevant depends on the experimental question being asked, particularly when also involving different extents of optogenetic manipulation.
Briefly, optogenetic manipulation requires the expression of light sensitive ion-selective channels (opsins) in targeted cells that, in the presence of light at a specific wavelength, depolarise (activate) or hyperpolarise (inhibit) the opsin-expressing cells. Light delivery techniques vary and are a determinant factor of the magnitude of the optogenetic manipulation. Indeed, one-photon field optogenetics is routinely used to target large cortical and subcortical areas, leading to the activity manipulation of large populations of opsin-expressing cells. Contrarily, single-cell targeted optogenetics (spiral scan) and holographic stimulation (using spatial light modulators) allow the manipulation of spatially restricted single neurons or ensembles of neurons. Knowing the limitations of these light delivery approaches is central to successfully achieve a well-designed optogenetic experiment, which I will discuss later.
During my PhD I used both one-photon field optogenetics and two-photon spiral-scan single cell targeted optogenetics. Consider the activation of a single neuron using single-cell targeted optogenetics as the addition of a few random notes from a single musician within a big orchestra playing loud fast-paced melodies. From a general audience perspective, those notes will probably pass by unnoticed, together with a few mistakes here and there from the other musicians, not changing the overall quality of the performance. To the musicians surrounding the intentionally mistaken one, however, the reaction to the off-key/off-beat notes may vary, but such notes will most certainly be noticed. Hence, when assessing whether single-cell optogenetic activation changes network activity using all-optical approaches, one should adjust the experimental methodology to the perspective of interest – either the nearby fellow musicians or the audience’s point-of-view.

Figure 2 - Example of reliable stimulation of a single opsin-expressing neuron using single-cell targeted spiral-scan optogenetics. ChRmine-mScarlet and jGCaMP7f coexpressing targeted neuron, with dF/F traces of the same neuron upon spiral-scan activation within the same imaging session.
In practical terms, if one is interested in understanding how precisely the activation of a single neuron impacts the activity of close by neurons, maybe a smaller field-of-view, with higher sampling rates, is the best experimental approach, such that fewer fast events are lost, and the data more closely represents what is happening within a more contained space. Contrarily, if one is interested in how single-cell manipulation affects the overall dynamics of the network in a large cortical area – the audience’s perspective – a larger FOV is required, at the cost of faster events’ detection and precision. Here, other aspects overwhelmingly dictate the overall dynamic of cortical activity, bringing me to another issue that dominated my PhD – movement.
How movement related neural representations dominate cortical activity?
Cortical activity in rodents, particularly in sensory areas, is known to be tightly correlated with movement (Vinck et al., 2015), such that spontaneous recordings of neural activity in primarily visual areas can be almost entirely explained by small face movements (Syeda et al., 2024), running traces and pupil dilation (a proxy for arousal state). Because in a large population of neurons these variables strongly dictate cortical activity, smaller less-noticeable events may be imperceptible when analysing two-photon calcium imaging data in awake mice.
Therefore, one must always account for both movement and a proxy for the brain’s internal state (e.g. pupil dilation in rodents) when analysis two-photon calcium imaging data in awake animals. Indeed, recent studies suggest that the same optogenetic perturbation during distinct states of engagement in a behavioural task, leads to different neural network and behavioural perturbations. This means that heterogeneity within optogenetic experiments (coupled or not with imaging) may be explained by contributions from neural representations associated with movement and/or the brain’s internal state. As such, face and eye cameras with high frame rate acquisition should be an integral part of any two-photon in vivo calcium imaging and/or behavioural set-ups, as information from these videos may be used to account for movement-related and arousal-dependent neural representation within the collected data. Movement and neural representations of the brains internal state are then integral part of the polyphony being played, as much as volume within an orchestra can mask a few off-key/off-beat notes here and there.
While working on my PhD project, I was interested in studying the effect of single cell optogenetic manipulation on cortical activity during postnatal brain development in mouse pups, and for several months we found no significant effect on overall cortical activity. It was only when we accounted for movement-related cortical representations that the real effect was observed and became significant.

Figure 3 - Body and face cameras are essential for controlling for movement-dependent neural representations when recording brain activity. These videos can be used to decode movement related neuronal representations in cortical regions and subsequently be used to eliminate its dominance of cortical activity dynamics.
Why not fight forte with fortissimo? A potential light-artefact problem.
Let’s now assume that an orchestra plays at full power with great volume, it may be harder for both the set of musicians surrounding a single mistaken one and the audience to spot single-mistakes here and there and, most certainly, everyone will leave with the same overall appreciation for the music played. However, if a large subset of musicians regularly and collectively makes mistakes – and do it loudly –, these more easily become noticeable, and both their fellow musicians and the audience may react with discontentment. This large perturbation is usually achieved with single photon field optogenetics, the focus of this section.
By field optogenetics I mean the illumination of a particular volume of tissue containing opsin-expressing cells without single-cell resolution, usually using one-photon visible light. Like a conductor can manipulate the volume of an ensemble within the orchestra without having to appoint each musician individually.
In this case, the magnitude of activity manipulation of opsin expressing cells it tightly related to light intensity and opsin expression levels in targeted neurons. Indeed, one usually assumes that maxing out optogenetic light power is the best way for optimal optogenetic effects. Although there is truth in this, as the presence of more energy from wavelength-specific photons increases the probability of conformational changes within the light-sensitive ion channels, leading to the production of large photocurrents, this raises a single issue that is sometimes overlooked, and it is critical in all-optical experimental approaches – light-artefacts.
These light-artefacts are not imaging related artefacts, such as bleed through or light contamination, as these can be fixed by removing contaminated frames or by hardware tricks, such as blanking the trigger signals. Here, I refer to cortical representations of the optogenetic light across superficial sensory cortical areas. Indeed, when I started my PhD, the general agreement was that mice cannot see red light, based on the absence of red cones in their retina (Peirson et al., 2018). This meant that the use of red-light sensitive opsins should not produce light-dependent neural representations, and changes in neural dynamics in optogenetic experiments could only be an effect of opsin expressing cells – this is not the case. After a few months of optimism, I finally decided to run light controls, naively because I would still notice drastic light modulatory effects on cortical activity when low to none expression levels of the opsin could be observed. To everyone’s surprise, I found that even at low optogenetic light powers (~1.8 mW) neurons responded to red-light (637 nm) both in visual and somatosensory areas in mice. This means that for every optogenetics experiment (coupled or uncoupled to imaging) one must find the right amount of light power that does not interfere with the brain’s internal state and/or neural representations of sensory stimuli, by promptly running the right light control experiments.
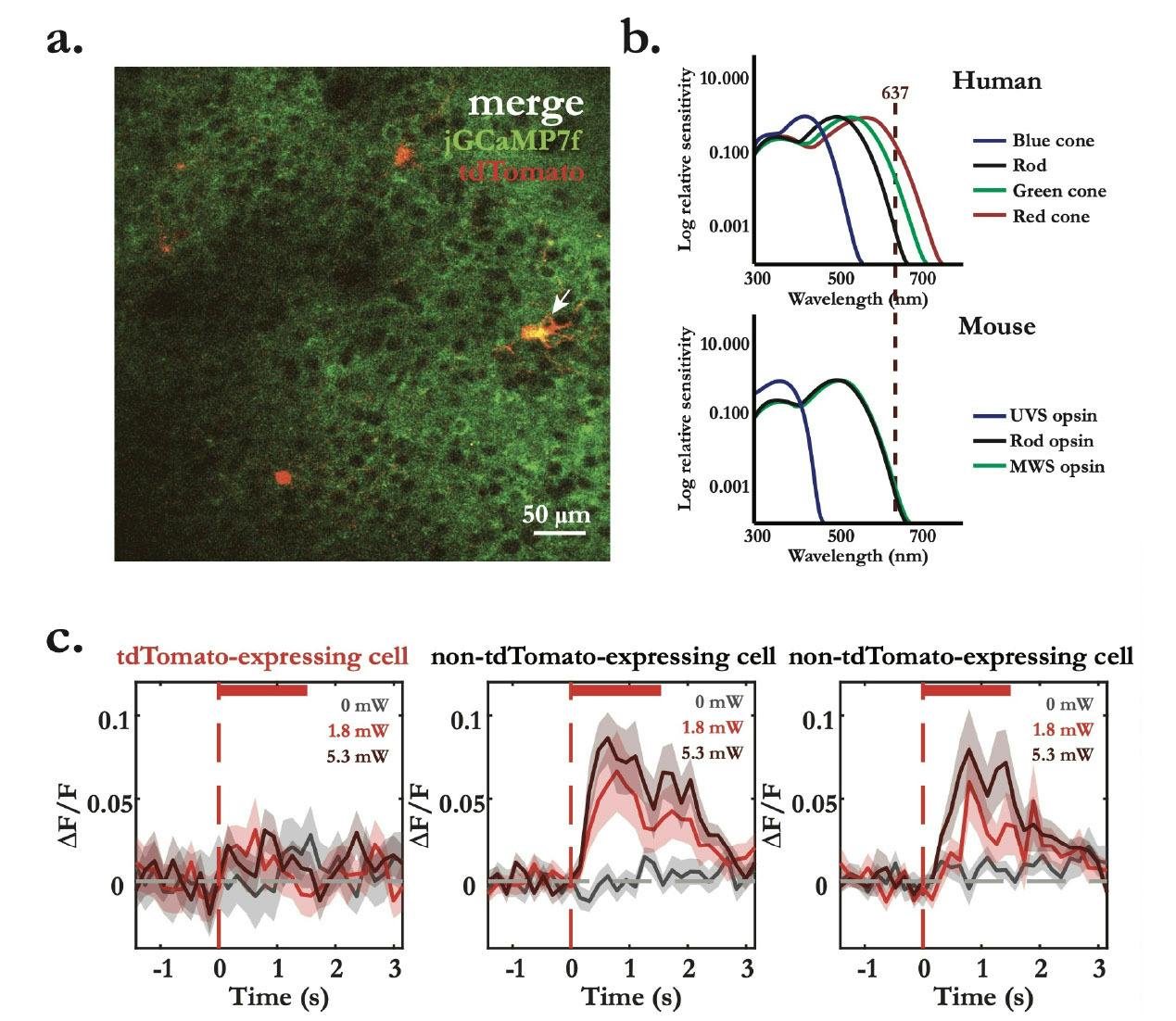
Figure 4 – Light control experiment shows optogenetic light-evoked calcium responses within photostimulation interval. a. Merge of plane 1 of L2/3 multiplane two-photon dual-channel recording in V1, showing the broad expression of jGCaMP7f (green) and targeted expression of tdTomato (red) in targeted neurons. b. Spectral of relative sensitivity of human and mouse visual pigments (adapted from Peirson et al., 2018). Red vertical dashed line represents the wavelength of the excitation laser used experimentally. c. Optogenetics light-evoked calcium mean responses (dF/F ± standard error of the mean) aligned to the onset of the stimulation laser (red vertical dashed line) with incresing laser powers (0 mW, 1.8 mW and 5.3 mW). tdTomato- labelled cell (left) shows no clean light evoked-responses, while two unlabelled cells (middle and right) show light-artefact calcium responses. Response lines represent the mean response of an average of approximately 30 trials in each condition.
One must mention, however, that light intensity needed to manipulate neural activity intimately depends on the expression levels of the opsin, light sensitivity and magnitude of induced photocurrents and the sub-cellular targeting of the opsin. Indeed, Antinucci et al., (2020) showed that variations in opsin efficiency is associated with its expression levels in zebra fish, where low opsin expression cannot induce strong enough photocurrents to depolarise or hyperpolarise the targeted cells. In this case, two approaches can be considered to tackle this issue: one can either try and increase expression levels of the opsin or develop opsins with are extremely sensitive to light, meaning that short, low-intensity light pulses are needed to induce huge photocurrents. Indeed, in the last few years multiple opsins have been engineered to produce larger photocurrents in the presence of low-intensity far-red and infrared light, namely ChrimsonR and ChRmine. Therefore, one must be careful when choosing the right opsin and the right light delivery approach to eliminate light-artefacts and efficiently and reliably photostimulate targeted neurons.
Finally, increasing protein expression levels and restrict expression to subcellular compartments are alternate ways to achieve strong and reliable manipulation of opsin-expressing neurons. This depends entirely on the use of genetic tools and viral vector technologies. Indeed, the easiest way to achieve transgenic expression involves either maintaining a strain of genetically modified animals that express the desired optogenetic tool or by crossing two strains. The latter requires the use of a ‘driver’ line, engineered to express a recombinase or transcription regulator such as Cre or Flpo (in rodents) in a specific cell population, along with an animal strain carrying a conditional gene controlled by the relevant driver. This approach, however, has been shown to produce weak and not uniform expression across all neuron subtypes (Madisen et al., 2012), and although collective efforts have been put into improving protein expression levels (Madisen et al., 2015), the creation of specific mouse lines is expensive and lacks the versatility and cost-efficiency needed, found in viral vector-based approaches.
Therefore, adeno-associated virus (AAV) and lentiviral vectors are the preferred tools to carry genes encoding optogenetic tools. These can beadministered directly into the brain tissue or through systemic injection to target specific circuits or broader brain regions, respectively (Challis et al., 2009). These viral approaches are commonly known to yield greater protein expression levels as multiple copies of the genes are transfected into all neurons in the injected area and, when under the control of transcription regulators like Cre and Flpo, should theoretically only be present in subtypes of neurons expressing either or both enzymes. These genetic tools (both transgenic line and viral-vector technologies) are widely used in neuroscience research and most of the time their efficacy and specificity are unquestioned – we just assume they work!
But I am writing this article, and by now you should guess what I am saying next – they do not work! Well, at least not always.
Check if your genetic tools truly label the cells you are interested in experimentally.
During my PhD I spent almost a year activating single interneurons, which expressed an opsin through a viral approach combined with a Crerecombinase mouse line and measuring the impact of their activation on the activity of the surrounding neurons using two-photon calcium imaging and single-cell spiral-scan two-photon optogenetics. The cells I was interested in were sparse and difficult to find, and I soon started wondering whether I could confirm that I was, for sure, stimulating the interneuron subtype I was interested in studying. It turned out that post hoc immunostaining and a few electrophysiology experiments later confirmed that most of the cells I was stimulating were not my interneurons of interest but pyramidal neurons. Indeed, Müller-Komorowska et al. (2020) highlighted the presence of nonspecific expression in limited excitatory cell populations in interneurontargeting Cre- driver lines, indicating that careful consideration is needed to minimise potential off-target effects. Similarly, Cheong et al. (2015) discussed the importance of comparative analysis of potential broad-spectrum neuronal Cre drivers, suggesting the need for comprehensive evaluation of
Cre-driver lines for their specificity and efficiency. Despite these finding, most people just assume that the tools always work. Hence, I suggest that before doing any imaging or behavioural experiments using optogenetics, one should always start by testing whether opsin-expression specificity is achieved by titrating viral concentrations and performing at least post hoc immunostaining for confirmation.
When available, use soma-targeted opsins, for photo-modulation of neuronal activity.
Finally, soma-targeted opsin expression has been shown to offer advantages in limiting non-targeted effects of photostimulation and achieve precise control of neural activity. Mardinly et al. (2018) engeneered somatargeted optogenetic tools such as ST- ChromeME and IRES-ST-eGtACR1, for optimised two-photon activation and suppression, respectively. Additionally, blue light-sensitive soma-targeted opsins (stCoChR) have also been shown to yield reliable photo-activation of targeted cells with 10-fold lower average power of stimulation light, contributing to the elimination of crosstalk with the imaging system (Forli et al., 2021).
These advances offer great benefits to achieve precise control of neural activity with low-light intensity, avoiding most light-related issues. Indeed, single-photon optogenetic stimulation combined with two-photon calcium imaging seems to be slowly replaced by holographic stimulation which uses infrared light and allows precise modulation of dozens of neurons simultaneously. These systems are, however, extremely expensive and much harder to operate.
Take home message: control, be sure, be sure again, have fun.
If you are new to all-optical approaches, welcome to the family! I hope this article gives you a general idea of the most important things to look out and control for when you are starting your experiments. I wish I had found something like this when I started my PhD. It really would have saved me a lot of time and tears.
I would suggest you think thoroughly about your research problem, and even question whether this all-optical approach is the best option available with the right temporal resolution you need to address what you are trying to answer. If so, think about potential light-artefacts and movement related neuronal representations in your data – it might not impact the outcome of your experiment, but it is better to be safe than sorry. If you are using genetic tools make sure they work for your experiment, even if everyone around you tells you they do – always check! More advanced optic tools are released every other month usually because they try to fix issues that are known to exist but not talked about, keep an eye out for those tools has someone might have just fixed your problem. Lastly, have fun with it! Once you trust your system and can control for any artifacts, your imagination is the limit. Be the conductor of this beautiful melody we are trying to understand.
About Filipe Machado Ferreira
Filipe has a BSc in Biochemistry from the University of Lisbon (Portugal, 2015) and has recently finished his PhD using all optical-approaches at the Centre for Developmental Neurobiology (CDN) King's College London, under the supervision of both Dr Adil Khan and Professor Juan Burrone. He is currently working as a research associate in the Khan lab, and his work focuses on combining two-photon calcium imaging, single-cell spiral-scan and holographic stimulation to understand complex levels of cognition at a microcircuit level involving different subtypes of interneurons.
References
Antinucci, P., Dumitrescu, A. S., Deleuze, C., Morley, H. J., Leung, K., Hagley, T., Kubo, F., Baier, H., Bianco, I. H., & Wyart, C. (2020). A calibrated optogenetic toolbox of stable zebrafish opsin lines (p. 2020.01.13.904185). bioRxiv. https://doi.org/10.1101/2020.0...
Challis, R. C., Ravindra Kumar, S., Chan, K. Y., Challis, C., Beadle, K., Jang, M.J., Kim, H. M., Rajendran, P. S., Tompkins, J. D., Shivkumar, K., Deverman, B. E., & Gradinaru, V. (2019). Systemic AAV vectors for widespread and targeted gene delivery in rodents. Nature Protocols, 14(2), 379–414. https://doi.org/10.1038/s41596...
Cheong, Soon K., Wenjun Xiong, Jennifer M. Strazzeri, Constance L. Cepko, David R. Williams, and William H. Merigan. ‘In Vivo Functional Imaging of Retinal Neurons Using Red and Green Fluorescent Calcium Indicators’. In Retinal Degenerative Diseases, edited by John D. Ash, Robert E. Anderson, Matthew M. LaVail, Catherine Bowes Rickman, Joe G. Hollyfield, and Christian Grimm, 135–44. Cham: Springer International Publishing, 2018. https://doi.org/10.1007/978-3-....
Forli, A., Pisoni, M., Printz, Y., Yizhar, O., & Fellin, T. (2021). Optogenetic strategies for high-efficiency all-optical interrogation using blue-lightsensitive opsins. eLife, 10, e63359. https://doi.org/10.7554/eLife....
Müller, C., & Remy, S. (2014). Dendritic inhibition mediated by O-LM and bistratified interneurons in the hippocampus. Frontiers in Synaptic Neuroscience, 6. https://www.frontiersin.org/ar...
Peirson, S. N., Brown, L. A., Pothecary, C. A., Benson, L. A., & Fisk, A. S. (2018). Light and the laboratory mouse. Journal of Neuroscience Methods, 300, 26– 36. https://doi.org/10.1016/j.jneu...
Russell, Lloyd E., Mehmet Fişek, Zidan Yang, Lynn Pei Tan, Adam M. Packer, Henry W. P. Dalgleish, Selmaan N. Chettih, Christopher D. Harvey, and Michael Häusser. ‘The Influence of Cortical Activity on Perception Depends on Behavioral State and Sensory Context’. Nature Communications 15, no. 1 (19 March 2024): 2456. https://doi.org/10.1038/s41467....
Syeda, A., Zhong, L., Tung, R., Long, W., Pachitariu, M., & Stringer, C. (2024). Facemap: A framework for modeling neural activity based on orofacial tracking. Nature Neuroscience, 27(1), Article 1. https://doi.org/10.1038/s41593...
Vinck, M., Batista-Brito, R., Knoblich, U., & Cardin, J. A. (2015). Arousal and locomotion make distinct contributions to cortical activity patterns and visual encoding. Neuron, 86(3), 740–754. https://doi.org/10.1016/j.neur...
Madisen, L., Garner, A. R., Shimaoka, D., Chuong, A. S., Klapoetke, N. C., Li, L.,van der Bourg, A., Niino, Y., Egolf, L., Monetti, C., Gu, H., Mills, M., Cheng,A., Tasic, B., Nguyen, T. N., Sunkin, S. M., Benucci, A., Nagy, A., Miyawaki, A., ... Zeng, H. (2015). Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron, 85(5), 942– 958. https://doi.org/10.1016/j.neur...
Madisen, L., Mao, T., Koch, H., Zhuo, J., Berenyi, A., Fujisawa, S., Hsu, Y.-W. A., Garcia, A. J., Gu, X., Zanella, S., Kidney, J., Gu, H., Mao, Y., Hooks, B. M., Boyden, E. S., Buzsáki, G., Ramirez, J. M., Jones, A. R., Svoboda, K., ... Zeng, H. (2012). A toolbox of Cre-dependent optogenetic transgenic mice for lightinduced activation and silencing. Nature Neuroscience, 15(5), 793–802. https://doi.org/10.1038/nn.307...
Mardinly, A. R., Oldenburg, I. A., Pégard, N. C., Sridharan, S., Lyall, E. H., Chesnov, K., Brohawn, S. G., Waller, L., & Adesnik, H. (2018). Precise multimodal optical control of neural ensemble activity. Nature Neuroscience, 21(6), Article 6. https://doi.org/10.1038/s41593...




)